Bacterial Functional Genomics and Antibiotics
Group Leader(s): Dr Hee-Jeon Hong
About us
The development of resistance to existing antibiotics, coupled with a sustained decline in the success rate of the discovery of new ones, is currently leading to a point in the future where many infections could essentially be untreatable by the compounds that are available. A fundamental understanding of bacterial antibiotic tolerance and resistance mechanisms will be an important part of any future strategy aimed at solving the growing problems with treating microbial infections.
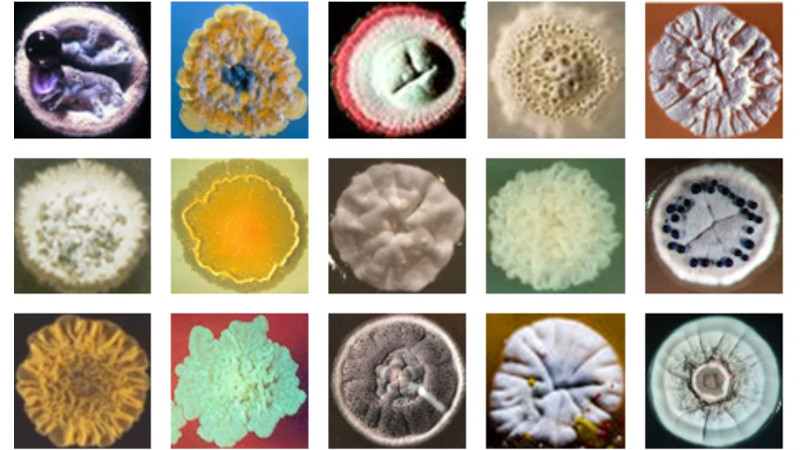
Our research involves using functional genomics approaches to characterise the response when growing cultures of bacterial cells are challenged with antibiotics. We are using a harmless, soil derived Gram-positive bacteria, Actinomycetes, as a model system to investigate the mechanisms involved in sensing, responding and adapting to antibiotic attack. Actinomycetes produce about 70% of known antibiotics and are the ultimate source of most antibiotic resistance genes. As a consequence, they possess many genes involved in sensing and responding to antibiotics, and are therefore an ideal system to use for furthering the understanding of the processes involved.
Related courses
Research impact
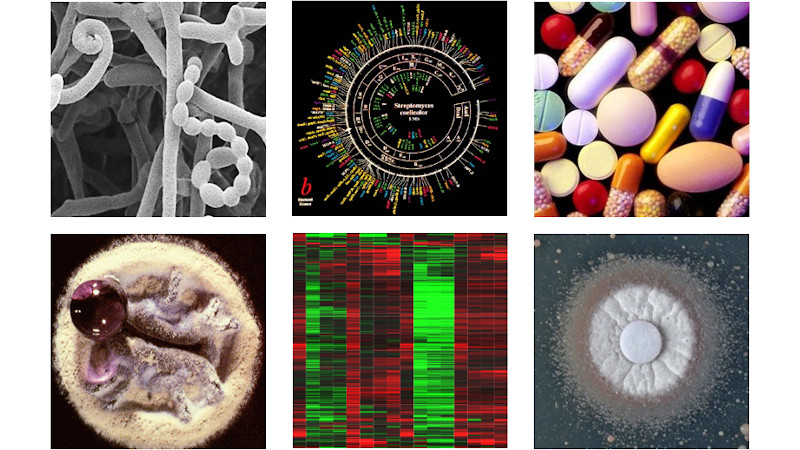
Understanding the genome-wide dynamics in response to antibiotics will extend our knowledge of the functions and systems that are important for bacterial antibiotic tolerance and resistance, and open ways by which these can be exploited in future antibiotic therapies.
We also aim to progress this research into pathogens responsible for hospital acquired infections. This work has direct implications for public health, contributing to efforts to understand the molecular basis of defensive responses and resistance to antibiotics in bacteria.
Leadership

Dr Hee-Jeon Hong
Senior Lecturer in Microbiology